Introduction¶
MM/PB(GB)SA method can be used for calculating binding free energies of non covalently bound complexes.
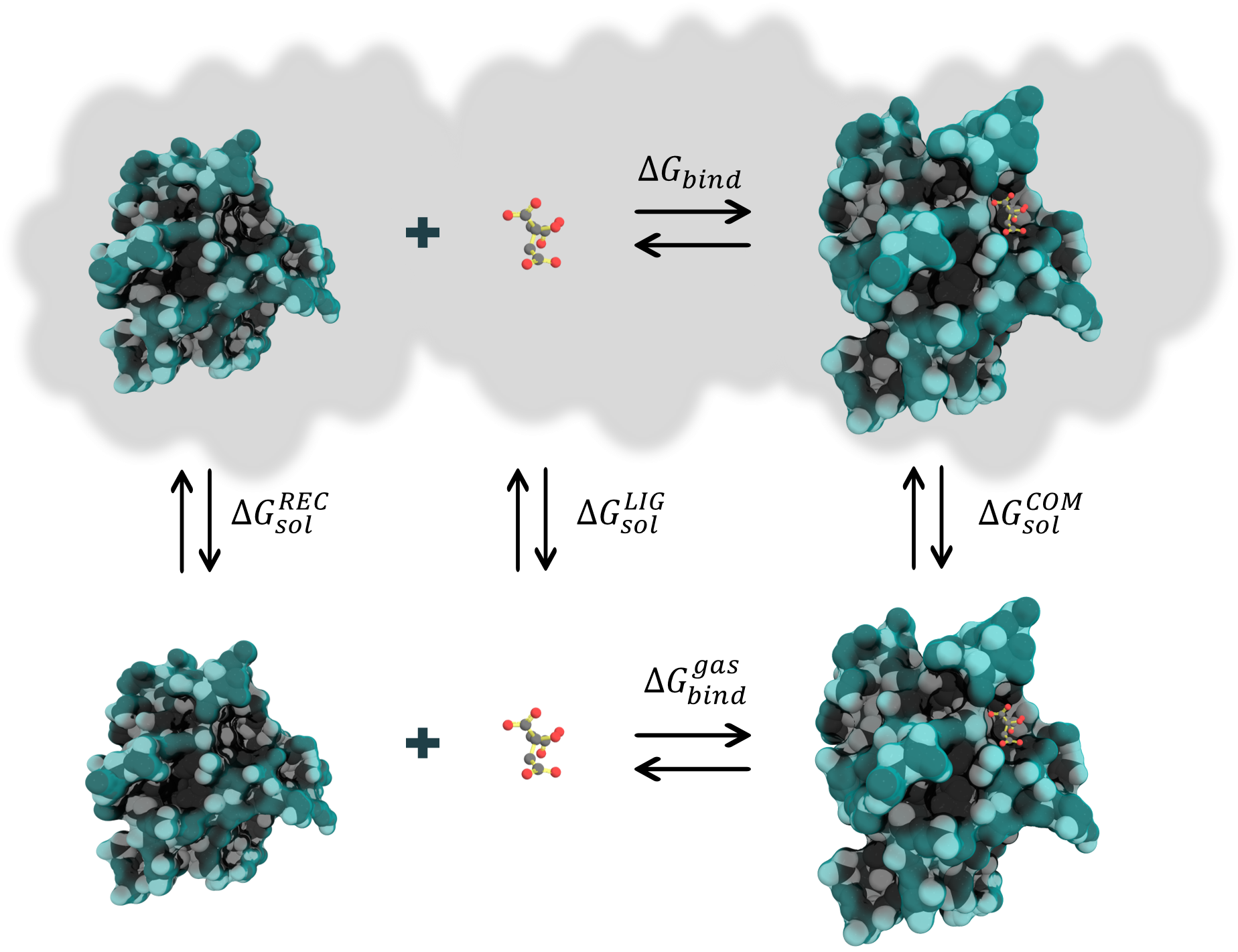
Figure 1. Thermodynamic cycle for binding free energy calculations
The free binding energy for a complex can be estimated as follows:
∆𝐺𝑏𝑖𝑛𝑑 = 〈𝐺𝐶𝑂𝑀〉−〈𝐺𝑅𝐸𝐶〉−〈𝐺𝐿𝐼𝐺〉
(1)
where each term to the right in the equation is given by:
〈𝐺𝑥〉 = 〈𝐸𝑀𝑀〉 + 〈𝐺𝑠𝑜𝑙〉 − 〈𝑇𝑆〉
(2)
In turn, ∆𝐺𝑏𝑖𝑛𝑑 can also be represented as:
∆𝐺𝑏𝑖𝑛𝑑 = ∆𝐻 − 𝑇∆𝑆
(3)
where ∆𝐻 corresponds to the enthalpy of binding and −𝑇∆𝑆 to the conformational entropy after ligand binding. When the entropic term is dismissed, the computed value is the effective free energy, which is usually sufficient for comparing relative binding free energies of related ligands.
The ∆𝐻 can be decomposed into different terms:
∆𝐻 = ∆𝐸𝑀𝑀 + ∆𝐺𝑠𝑜𝑙
(4)
where:
∆𝐸𝑀𝑀 = ∆𝐸𝑏𝑜𝑛𝑑𝑒𝑑 + ∆𝐸𝑛𝑜𝑛𝑏𝑜𝑛𝑑𝑒𝑑 = (∆𝐸𝑏𝑜𝑛𝑑 + ∆𝐸𝑎𝑛𝑔𝑙𝑒 + ∆𝐸𝑑𝑖ℎ𝑒𝑑𝑟𝑎𝑙) + (∆𝐸𝑒𝑙𝑒 + ∆𝐸𝑣𝑑𝑊)
(5)
The gas phase free energy contributions (∆𝐸𝑀𝑀) are calculated by sander within the AmberTools package according to the force field used in the MD simulation.
The ∆𝐺𝑠𝑜𝑙 is given by:
∆𝐺𝑠𝑜𝑙 = ∆𝐺𝑝𝑜𝑙 + ∆𝐺𝑛𝑜𝑛−𝑝𝑜𝑙 = ∆𝐺𝑃𝐵/𝐺𝐵 + ∆𝐺𝑛𝑜𝑛−𝑝𝑜𝑙
(6)
where:
∆𝐺𝑛𝑜𝑛−𝑝𝑜𝑙𝑎𝑟 = 𝑁𝑃𝑇𝐸𝑁𝑆𝐼𝑂𝑁 ∗ ∆𝑆𝐴𝑆𝐴 + 𝑁𝑃𝑂𝐹𝐹𝑆𝐸𝑇
(7)
or,
∆𝐺𝑛𝑜𝑛−𝑝𝑜𝑙 = ∆𝐺𝑑𝑖𝑠𝑝 + ∆𝐺𝑐𝑎𝑣𝑖𝑡𝑦 = ∆𝐺𝑑𝑖𝑠𝑝 + (𝐶𝐴𝑉𝐼𝑇𝑌𝑇𝐸𝑁𝑆𝐼𝑂𝑁 ∗ ∆𝑆𝐴𝑆𝐴 + 𝐶𝐴𝑉𝐼𝑇𝑌𝑂𝐹𝐹𝑆𝐸𝑇)
(8)
In the above equations, ∆𝐸𝑀𝑀 corresponds to the molecular mechanical energy changes in the gas phase. ∆𝐸𝑀𝑀 includes ∆𝐸𝑏𝑜𝑛𝑑𝑒𝑑, also known as internal energy, and ∆𝐸𝑛𝑜𝑛𝑏𝑜𝑛𝑑𝑒𝑑, corresponding to the van der Waals and electrostatic contributions. The solvation energy is determined differently, depending on the method employed. In the 3D-RISM model, both components -polar and non-polar- of the solvation energy are calculated. However, the PB and GB models estimate only the polar component of the solvation. The non-polar component is usually assumed to be proportional to the molecule's total solvent accessible surface area (SASA), with a proportionality constant derived from experimental solvation energies of small non-polar molecules (eq. 7). Alternatively, a modern approach that separates non-polar solvation free energies into cavity and dispersion terms can be used. In this approach, SASA is used to correlate the cavity term only, while a surface-integration method is employed to compute the dispersion term (eq. 8).
Furthermore, the entropic component is usually calculated by normal modes analysis (NMODE). The translational and rotational entropies can be estimated using standard statistical mechanical formulas. Nevertheless, calculating vibrational entropy using normal modes is computationally expensive because it requires expanding the internal coordinate covariance matrix for all degrees of freedom for a set of minimized structures. Conversely, the Quasi-harmonic (QH) approximation is less computationally expensive, although it requires a considerable number of frames to converge. Recently, other alternatives have been developed, such as NMODE in truncated systems, which considerably reduces the computational cost. Interaction Entropy (IE) is another novel method that calculates the entropic component of the binding free energy directly from MD simulations without any extra computational cost. This method is numerically reliable, more computationally efficient, and superior to the standard NMODE approach, as shown in an extensive study of over a dozen randomly selected protein-ligand binding systems.
Typically, two approaches are used for MM/PB(GB)SA calculations, known as Single Trajectory Protocol (STP) and Multiple Trajectory Protocol (MTP). In STP, both the receptor and the ligand trajectories are extracted from that of the complex. This approach is valid when the bound and unbound states of the receptor, and the ligand are similar. It is computationally less expensive than the MTP approach since only a simulation of the complex is required. Additionally, the potential internal terms (e.g., bonds, angles, and dihedrals) cancel exactly in STP since these terms are the same in both bound and unbound states. On the other hand, the MTP is a more realistic approach because it considers multiple trajectories (i.e., complex, receptor, and ligand). However, significant conformational changes can lead to numerous errors. In practice, a detailed study of the system is required to select the approach to be used.
Literature¶
Further information can be found in Amber manual:
- MMPBSA.py
- The Generalized Born/Surface Area Model
- PBSA
- Reference Interaction Site Model
- Generalized Born (GB) for QM/MM calculations
and the foundational papers:
as well as some reviews and expert opinions:
Created: February 8, 2021 07:10:13